The views expressed in this paper are those of the writer(s) and are not necessarily those of the ARJ Editor or Answers in Genesis.
Abstract
The melanocortin 1 receptor (MC1R) is located on the surface of melanocytes (pigment cells) and is involved with switching melanin synthesis from the lighter red to yellow pheomelanin to the darker brown to black eumelanin. Most animals are capable of producing both pigments and have various shades of color distributed throughout their hair coats. Mutations in the MC1R can add interesting color patterns ranging from the all black phenotype of Chinese Meishan pigs to the all red color of the Irish Setter. Given the number of alleles within baramins (created kinds), much of the diversity at this locus must have developed since the genetic bottleneck at the Flood where only a single breeding pair was preserved for most baramins. Similarly, humans carry more alleles than can be accounted for by Noah and his family. Since mutations at this locus are not known to directly cause any disease and they add to the beauty and variety we see today, it appears that this locus was designed to vary. Furthermore, since there are non-random patterns that have been observed which cannot be explained by known selection factors, it appears plausible that many of these mutations may actually be directed.
Keywords: Coat color, Directed mutations, MC1R, Melanocortin receptor, Mutations, Skin color, Hair color
Introduction
The historical account of Creation in Genesis relates that animals were created according to their kinds, with the ability to reproduce, and were given instructions (or blessed) to fill the earth and seas (Genesis 1:21–22, 24–25). However, no information is recorded about the number of individuals created within each kind. In contrast, the Bible does mention that two humans were created (Genesis 1:26–28; 2:8, 18–25; 3:20). Later, the Bible indicates that terrestrial and flying animals underwent a severe genetic bottleneck at the time of the Flood, estimated by Ussher to be 2348 BC (Genesis 6–8; Ussher 2003, pp. 19–21). The majority of the created kinds of terrestrial animals (that is, the unclean animals, which likely included all non-ruminants; Leviticus 11:1–8, 26–47) were represented by a single breeding pair on the Ark (Genesis 6:17–7:2). Humans also experienced a genetic bottleneck during the Flood when eight individuals were saved (Genesis 7:1, 7, 13; 1 Peter 3:20).
The creation model must be able to account for diversity observed today within created kinds (baramins) and the human population given this relatively recent bottleneck. Clearly animals have adapted to many different environmental conditions since the Flood. For example, there are foxes (genus Vulpes) which are adapted to arctic conditions (for example, V. lagopus) while others live in the desert (for example, V. zerda; Myers et al. 2008). There have been reports in the literature of rapid adaptation occurring in a variety of animals including lizards (Herrel et al. 2008), finches (Grant and Grant 2006), mosquitoes (Byrne and Nichols 1999), and guppies (Arendt and Reznick 2005). Rapid adaptation fits well within the creationist model since God is a provider who cares for His Creation (Psalm 147:8–9; Matthew 6:25–34) and intends the earth to be inhabited (Isaiah 45:18).
There is a need to more fully characterize the genetic basis of intrabaraminic (within kind) diversity to further develop the creationist model. Historically, many creationists have limited explanations of intrabaraminic diversity to initial variability, degenerative random mutations, and natural selection. Given that baraminologic investigation has found that holobaramins tend to include whole families or even several families of animals, there is serious question as to how well the above mechanisms account for what we observe in the world around us. For example, the family Felidae (cats) is believed to represent a holobaramin as is the superfamily Physeteroidea (grouping of toothed whales) and the suborder Mysticeti (baleen whales) (Wood 2006). Given the degree of variability within these taxa, creationists should be asking what kind of genomic differences exist within a holobaramin and if current explanations truly account for these differences.
The scientific literature is filled with studies evaluating the genetic basis of coloration in mammals and birds. There are a number of different loci known to influence color; some have been more fully investigated than others. Creationists have used the fact that there are several well known loci to attempt to explain variation in human skin (and hair) color using primarily Mendelian genetics (Parker 2006). While red hair is acknowledged to be the result of mutation, the variation in humans is presented as being primarily the result of Mendelian genetics and natural selection (Anonymous 1980; Moore 1996).
One locus commonly associated with color variation is the melanocortin 1 receptor (MC1R) locus. This provides a practical starting point to search for intrabaraminic and interbaraminic patterns in protein coding genes. Emphasis will be placed on unclean animals, since they were represented by a single breeding pair less than 4500 years ago.
Data from humans, who were represented by two individuals at creation and were reduced to eight individuals during the Flood, will be included as well.
The MC1R locus
The MC1R locus codes for a seven transmembrane (TM) G-protein coupled receptor (GPCR). This superfamily has over 1000 members which together account for more than 1% of mammalian genomes. The MC1R belongs to the class A, rhodopsin family, of GPCRs and is one of a five-member subfamily, the melanocortin (MC) receptors. Since receptors in this family and subfamily share many basic features, findings in one receptor may have implications for other receptors in the group (García-Borrón, Sánchez- Laorden, and Jiménez-Cervantes 2005).
The MC1R is expressed on the surface of melanocytes. There are two basic types of pigments produced by melanocytes. Pheomelanin is a red to yellow pigment; eumelanin is a darker, more photoprotective brown to black pigment. Activation of the MC1R by its ligand, primarily α-melanocyte stimulating hormone (α-MSH)1, switches biosynthesis from the default pathway for pheomelanin to production of eumelanin. Mammals commonly have a MC1R antagonist, the agouti protein, which competitively binds the MC1R preventing its ligand from binding and activating it. Thus, by varying levels of α-MSH and agouti protein, the wild type receptor can effectively switch melanin biosythesis between these two forms of melanin (García-Borrón, Sánchez-Laorden, and Jiménez- Cervantes 2005; Klungland and Våge 2000).
Humans normally respond to ultraviolet (UV) light by increasing production of the MC1R ligand. When the ligand binds the MC1R, there is activation of the G protein and stimulation of adenylyl cyclase. Through a series of biochemical reactions that follow, the melanocyte switches from pheomelanin to eumelanin biosynthesis; this protects the skin from many of the damaging effects of UV light. However, this pathway is impaired in individuals with red hair and fair skin with poor tanning ability (García-Borrón, Sánchez- Laorden, and Jiménez-Cervantes 2005).
Within Kind Variability
Pigs and wild boars (Sus scrofa)
The pig MC1R locus is found on chromosome 6p and codes for 320 amino acids. This is three amino acid residues longer than the MC1R of most other mammalian species examined to date.2 The extra length is due to an apparent tandem duplication of codons 29 through 31 in pigs (Kijas et al. 2001). Gustafsson et al. (2001) reported seven different MC1R alleles had been identified and were associated with various coat color patterns seen in swine.
Two alleles are associated with the wild type phenotype. The first; found in the European wild boar, is the reference sequence; the second, found in the Japanese wild boar, has a synonymous substitution. Thus, the wild type alleles code for the same amino acid sequence (Giuffra et al. 2000).
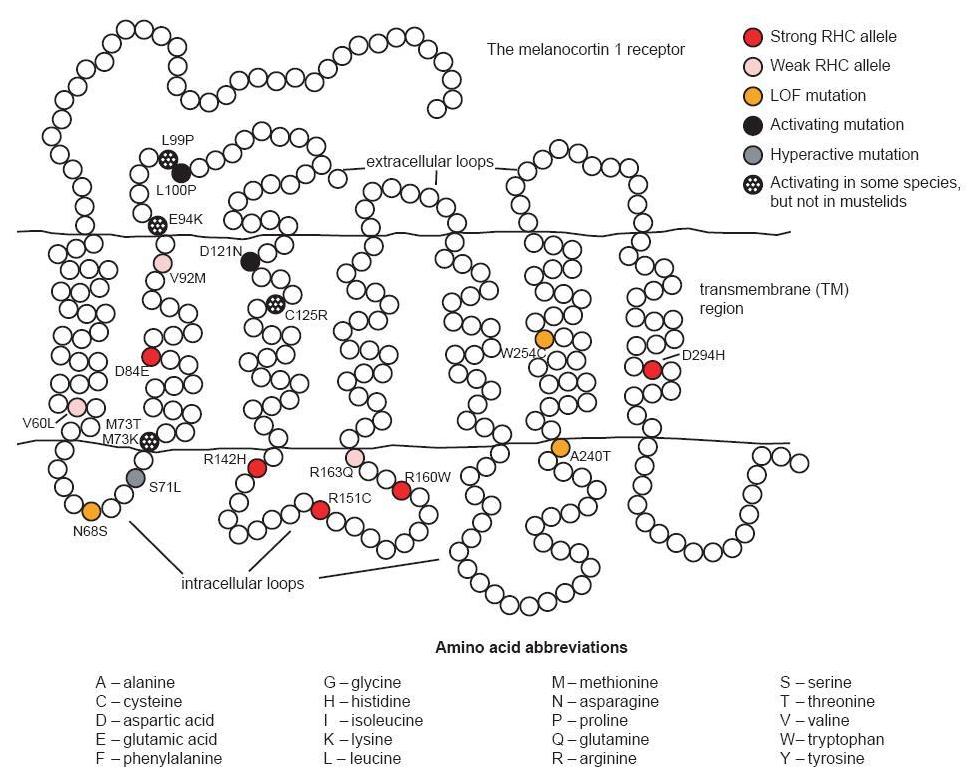
There are three alleles associated with a constitutively active receptor and black phenotype. The first of these is found in British Large Black and Chinese Meishan pigs.3 It contains two synonymous substitutions (one of which is identical to that found in the Japanese wild boar) and two nonsynonymous substitutions. The first amino acid change is V92M, a substitution also identified in red haired humans; this is not considered to significantly affect the phenotype in these pigs (see fig. 1). The second substitution, L99P, is identical to a mutation causing a dominant black phenotype in cattle and is likely the cause of the black phenotype in these pigs. A second allele associated with the black phenotype in Hampshire pigs involves a synonymous substitution at codon 17 and D121N (Gustafsson et al. 2001; Kijas et al. 1998).4 The identical mutation causing D121N was one of two nonsynonymous mutations in an allele associated with the dominant black color in several breeds of sheep (Våge et al. 2003). The third allele coding for a constitutively active receptor was identified in the Meishan and is identical to the first with an additional substitution, V122I. It is believed that this last mutation does not obviously affect the phenotype since isoleucine is present at this position in the wild type receptor of the horse and fox (Gustafsson et al. 2001).
One allele is associated with a loss of function mutation and the red phenotype in the Duroc. It codes for two amino acid substitutions: A161V and A240T. The first probably does not significantly affect the function of the receptor since this is a variable residue position and the functional horse receptor also carries a valine in this position. The second substitution occurs in TM6 which is highly conserved across six mammalian species and all five MC receptors. The substitution of a hydrophobic alanine with a polar threonine may alter the α-helical structure of the TM domain (Kijas et al. 1998). Several of the TM regions, including TM6, are believed to be important in forming an agonist binding pocket (García-Borrón, Sánchez-Laorden, and Jiménez-Cervantes 2005). Without this binding pocket, the MC1R ligand is unable to effect the switch from pheomelanin to eumelanin synthesis.
By far the most interesting allele is the one associated with the black spotting phenotype in Pietrain pigs. On initial investigation it appeared to be the same allele as the dominant black found in the Hampshire with D121N (Kijas et al. 1998). However, complete sequencing revealed that the allele also contains a 2-base pair (bp) insertion at codon 23 (nt67insCC). This results in a frameshift with a premature stop codon. The severely truncated protein is nonfunctional. The black spots were shown to be from somatic reversion which restores the reading frame and codes for a constitutively active receptor (Kijas et al. 2001).
Dogs (Canis familiaris) and foxes (Vulpes spp.)
All members of the family Canidae are considered monobaraminic (Wood 2006). Within this family the MC1R of dogs and several species of foxes have been sequenced. The gene codes for a 317 amino acid receptor. Newton et al. (2000) compared the nucleotide sequence from seven breeds and found six polymorphic sites. Two of these were synonymous changes. Two of the nonsynonymous changes, T105A and P159Q, may have no significant effect on receptor function. This is suspected because the first varied between animals of the same breed and in the case of the second, specific to the Doberman, it is not correlated with variant phenotypes and glutamine, Q, is found in this position in the chicken receptor.
There are two amino acid substitutions in the dog that may be related to phenotypic variation in coat color. The first is an R306ter mutation found in the Yellow Labrador, Golden Retriever, and Irish Setter. This truncated receptor appears non-functional and this loss of function mutation correlates perfectly with the yellow/red phenotype in all breeds tested with the exception of the Red Chow. Interestingly, the allele carried by the Yellow Labrador is distinct from that of the Golden Retriever and Irish Setter. This has led to speculation that the R306ter mutation has arisen twice, or undergone gene conversion (Newton et al. 2000).
The second substitution, S90G, may be associated with a constitutively active receptor and the black phenotype. It occurs in the second TM domain where other activating mutations have been known to occur. However, further biochemical studies will be necessary to establish the functional significance, if any, of this variant (Newton et al. 2000). Additionally, it has been known for years that the dominant black phenotype in dogs can be inherited in an unusual manner. Recent studies showed variation in the MC1R gene could not account for the phenotype in the black Labrador Retriever (Kerns et al. 2007). This led to the discovery that another protein, recently identified as ß-defensin, can act as a MC1R ligand and contribute to coat color in dogs (Candille et al. 2007).
The dog MC1R is 96% identical to that of the Arctic fox (Vulpes lagopus; Newton et al. 2000). A variant allele in the Arctic fox containing two mutations, G5C and F280C, was associated with the blue phenotype, which is darker and does not express the typical white winter coat. One or both of these mutations appear to result in a constitutively active receptor, although further biochemical studies are needed to clarify the underlying basis for this (Våge et al. 2005).
Våge et al. (1997) sequenced the MC1R from 17 red foxes (Vulpes vulpes) and found six polymorphic sites. Only one of these, C125R, correlated perfectly with the dominant dark Alaskan phenotype. Unlike examples of a constitutively active receptor in other animals, individuals carrying this mutation could express some pheomelanin based coloration depending on the allele carried at the agouti locus.
Cats (Felidae)
Previous baraminologic research suggested that the family Felidae is a holobaramin (cited in Wood 2006). The cat MC1R is also 317 amino acid residues in length. The black phenotype in the domestic cat (Felis catus) is a recessive trait and has been shown to be associated with a deletion mutation at a different (agouti) locus (Eizirik et al. 2003).
The dominant black phenotype in the jaguar (Panthera onca) is associated with an in frame15 bp deletion that eliminates amino acids 101 to 105. This allele also has two substitutions in one codon resulting in L106T immediately following the deletion. Nucleotide position 825 is T/C polymorphic in the wild type, but only T was found at this position in the allele carrying the deletion (Eizirik et al. 2003).
The semidominant melanistic phenotype in the jaguarundi (Herpailurus yaguarondi) is associated with a 24 bp in frame deletion that eliminates amino acids 95 through 102. This gene also carries nonsynonymous substitutions, P22L, I63V, and Q310R. The first two are conservative in nature. Additionally, cattle carry leucine at position 22 in the wild type receptor; humans and mice have a similar amino acid at 310, lysine. Therefore the authors suggest that the amino acid substitutions likely have less impact on the phenotype than the deletion (Eizirik et al. 2003).
The deletion mutations in the domestic cat, jaguar, and jaguarundi appear to be species specific and have not been detected in melanistic individuals from five other felid species tested: leopard (Panthera pardus), Geoffroy’s cat (Oncifelis geoffroyi), oncilla (Leopardus tigrinus), pampas cat (Lynchailurus colocolo), and Asian golden cat (Catopuma temmincki; Eizirik et al. 2003).
The weasel family (Mustelidae)
While the baraminic status of this family has not been determined, interesting patterns have been found in the MC1R gene within Mustelidae. Hosoda et al. (2005) examined the 5’ flanking region and beginning MC1R sequence in 17 mustelid species from four genera. Some species exhibited intraspecies variation. For example the yellow-throated marten (Martes flavigula), sable (M. zibellina), and stone marten (M. foina) carried one, two, and three polymorphic sites within the coding region respectively. In contrast, five individual American pine martens (M. martes) carried identical sequences. Overall, the mustelids had a high proportion of nonsynonymous single nucleotide polymorphism (SNP). Within the Martes/Gulo clade, 23 of the 29 SNPs were associated with an amino acid substitution; all but one of the intraspecific SNPs were nonsynonymous. Within the Mustela clade, 21 of the 31 SNPs were nonsynonymous.
Several amino acid changes observed in the mustelids are associated with a constitutively active receptor in other animals, yet surprisingly no melanistic phenotype was observed. Six nonmelanistic Mustela individuals carry E94K, which corresponds to the E92K associated with the black phenotype in the mouse. M. martes carried C125R, which is identical to the silver allele in the red fox, yet there was no substantial similarity in coat color. A non-melanistic American mink (Mustela vision) carried L99P associated with the black phenotype in cattle and pigs. M73T, which is found in the black chicken and is in the same location as the M73K in black sheep, was found in the non-melanisitc badger (Meles meles; Hosoda et al. 2005; Våge et al. 2003).
Mutations associated with loss of function followed a more expected pattern. The sable (M. zibellina) normally has a dark brown phenotype. An individual with a rare light yellow coat was homozygous for C35F and N68S. A gray brown sable was heterozygous at the 68 position (Hosoda et al. 2005).
Four independent indel events were identified in the Martes/Gulo clade. First, a 15 bp deletion affecting TM2 (codons 94-98) was identified in G. gulo. Second, a 45 bp in frame deletion (codons 98-112) was identified in M. martes, M. americana, M. melamous, and M. zebellina species. The last two indels were found in the stone marten (M. foina): a 10 bp duplication followed closely by a 28 bp deletion. While, deletions in this region have been associated with melanism in jaguars and jaguarundis as described above, the indels described in these mustelids are not associated with melanistic phenotypes (Hosoda et al. 2005).
Mice (Mus musculus)
The MC1R locus in the mouse is at the distal end of chromosome 8 and codes for a protein only 315 amino acid residues in length (García-Borrón, Sánchez- Laorden, and Jiménez-Cervantes 2005; Klungland and Våge 2000). There are three natural dominant MC1R mutations that have been identified in mice. The sombre and sombre-3J alleles code for E92K and L98P respectively. The mutations are located in the outer portion of TM2; both result in a constitutively active receptor and black phenotype. The tobacco allele, S69L, results in a hyper-active receptor and mice with a black dorsum and agouti flanks (García- Borrón, Sánchez-Laorden, and Jiménez-Cervantes 2005; Wada et al. 2005).
Two recessive alleles have been identified in mice. The recessive yellow involves the deletion of nucleotide 549 resulting in a frame shift and premature stop 12 codons later. The tawny phenotype is dominant to the recessive yellow and is characterized by a reduced number of black hairs. This allele carries five SNPs compared to the reference sequence; two silent, V101A, V216A, and W252C. The first four are identical to those found in wild type individuals in the population in Japan. Tryptophan, W, is conserved at position 252 in all reported animals and is part of TM6. This non-conservative substitution likely changes the α-helical structure of the TM domain and quite possibly reduces ligand binding affinity (García- Borrón, Sánchez-Laorden, and Jiménez-Cervantes 2005; Wada et al. 2005).
Humans
The human MC1R is found on chromosome 16q (Harding et al. 2000). More than 60 non-conservative variants have been identified (García-Borrón, Sánchez- Laorden, and Jiménez-Cervantes 2005). Some of these variants are associated with fair skin and red hair color (RHC). Strong RHC alleles include the common R151C, R160W and D294H, and the less common D84E and R142H. Weaker RHC alleles include V60L, V92M, and R163Q. A number of the other allelic variants may be associated with some degree of loss of function too. Several mechanisms related to loss of function have been identified: decreased ability to stimulate cAMP production from impaired G-protein coupling, retention of MC1R intracellularly resulting in decreased cell surface expression, and decreased affinity of the receptor for its MC ligand (García- Borrón, Sánchez-Laorden, and Jiménez-Cervantes 2005). Additionally, the strong RHC alleles have been shown in vitro to exert a dominant negative effect on wild type receptors (Beaumont et al. 2007).
Variant alleles have been associated with an increased risk of various skin cancers including melanoma. Cancer risk varies depending on which variant alleles are carried. Some of the risk appears to be independent of skin type or hair color, suggesting that the MC1R affects risk through one or more non-pigmentary pathways as well (Bastiaens et al. 2001; Box et. al. 2001; Fargnoli et. al. 2006; Kennedy et al. 2001; Raimondi et al. 2008). There are a number of other factors which affect the risk of melanoma including sun exposure and variants in other genes (Bennett 2008; Fargnoli et al. 2008; Walsh 1999).
Harding et al. (2000) examined MC1R polymorphisms in European, Asian, and African human populations. Five haplotypes were identified in Africans (who were from The Gambia and Ivory Coast), but all substitutions were in silent, third base pair positions. They concluded:
The absence of amino acid variants in Africa—as well as their low frequency in African Americans and in Asians from Papua New Guinea and India …, where skin pigmentation is typically very dark— implies strong functional constraint on MC1R, probably as a means to minimize sensitivity to UV radiation. (Harding et al. 2000, p. 1354)
Harding et al. (2000) begin with the assumption that mutations are relatively random, chance copying errors. Using various statistical tests they infer strong selection within the African population and attribute the incredible diversity of MC1R in Europeans to relaxed functional constraints. Yet, if the populations sampled give a fairly accurate representation of MC1R diversity,5 there is another viable alternative that should be considered.
It is true that in humans variant MC1R proteins are often associated with some degree of loss of function. Some of these variants have been shown to be associated with an increased risk of various skin cancers, including malignant melanoma. Certainly the equatorial regions of earth have greater sun exposure which would further increase cancer risk. However, the strong selection suggested by Harding et al. (2000) appears much stronger than these factors can account for. For example, melanoma rarely strikes before the childbearing years. Lachiewicz et al. (2008) found the mean age of onset at 57 years. The peak frequency was 54 years for early onset and 74 years for late onset melanoma. Furthermore, only strong RHC alleles are known to significantly affect MC1R function in the heterozygous condition. Given this, selection against variants would be expected to be quite weak and primarily directed toward individuals that carry two copies of a single variant, a copy of two different variants, or a strong RHC variant. So, despite the fact that untreated melanoma is deadly, increased risk of skin cancer should not provide sufficient selection to cause the MC1R protein to be invariant in African populations. It is possible that mutation rate in the MC1R gene is affected by epigenetic factors such as sun exposure. It has been suggested that variant alleles may be helpful at higher latitudes to allow for more effective vitamin D synthesis and prevent rickets. Perhaps there are underlying mechanisms that allow for mutation rate in this gene to be influenced by environmental sun exposure. Such mechanisms may also be able to bias the location of the mutations.
Between Kind Variability
Comparisons across different taxa are often alluded to in the various articles mentioned here. Such comparisons have been helpful in elucidating various details about MC1R structure and function in general, although certain details remain taxon specific. It is interesting to note that certain identical mutations appear in different baramins. The L99P (CTG→CCG) in cattle and pigs as well as the D121N (GAC→AAC) in sheep and pigs are associated with constitutively active receptors and a black phenotype (Klungland and Våge 2000). Perhaps there are a limited number of ways to produce a constitutively active receptor and these black phenotypes have been selected by man. Yet surprisingly, L99P (CTG→CCG) was found in a non-melanistic American mink (Mustela vison, GenBank: AB189816, nucleotides 295-297). More detailed examination of rapidly accumulating data on the MC1R in various species will be necessary to determine if there is significant bias in the location mutations occur in animals and if selection can truly account for the bias.
Hosoda et al. (2005) made several interesting observations with regards to deletion events. In two of the three mustelid deletions, hexanucleotide direct repeats were observed flanking the deletion; this is similar to the pattern found in the jaguarundi. Other observations involving mustelid deletions were the deletion of a repeated triplet at the 3’ end of the deletion and the presence of a short palindromic region near the center of the deleted segment. This is significant because repeats have generally been considered a sign of degeneration in the genome. These observations imply that repeats may act to format the genome for future indel mutations (and perhaps other mutations as well). The bias toward in frame indels further supports this. Although the mustelid deletions occur in the same region as deletions implicated in melanism in the jaguarundi, golden-headed lion tamarin, and jaguar, there is no melanistic pattern found in these mustelids. This, and the fact that LOF mutations are not lethal, argues against natural selection being a major factor in the observed pattern of in frame indels.
Conclusions
Intrabaraminic diversity clearly indicates that allelic diversity has increased since the time of the Flood when a maximum of four alleles would be expected to have been preserved in the unclean animals represented by a single pair on the Ark. The variability in this locus accounts for much of the variability and beauty in coat colors normally seen in animals today. Humans, who have been surveyed in more detail, also show an incredible amount of diversity at this locus that must have arisen since the Flood. No disease is known to be caused by variation in this gene, although some variants may confer an increased risk of certain diseases. Still, the development of these diseases is dependent on other factors. From a creationist viewpoint, the MC1R locus appears to be designed to vary.
These data suggest that at least some mutations occurring at this locus may be directed to some degree. First, there are several identical activating mutations in different baramins. It has been suggested that chance errors in DNA replication were fixed in the population by selection for the black phenotype, which can only occur in a limited number of ways. Yet this is challenged by the fact that among the various mustelids four amino acid substitutions and several deletion mutations normally associated with activating mutations in other taxa have been identified, yet they have no black phenotype which can be selected. It remains unclear if these mutations in mustelids have any effect on a selectable phenotype. Secondly, the bias toward nonsynonymous substitutions in the mustelids is quite curious; there is no obvious selection that accounts for this pattern. Third, there are a surprising number of in-frame deletions in both the felids and the mustelids. Most indels would be expected to cause a frame shift and generally result in a premature stop codon and a red to yellow phenotype, as in the red Duroc and Golden Retriever. So, again, a nonrandom pattern of mutations is not associated with any known selection pressure that can account for it. Finally, and by far the most powerful argument because it has been studied in greater detail, the pattern of human variants cannot be explained by known selection forces. Thus, the MC1R appears to be a genetic locus that was designed to change and directed mutations are quite plausibly involved. More detailed studies across baramins with appropriate statistical analysis should allow for stronger conclusions.
It will be interesting to see if other loci affecting coat color are as amenable to mutations as the MC1R. It is quite possible that other receptors, particular other GPCRs, may provide a strategic place to look for loci amenable to mutational change and directed mutations. Patterns in both SNPs and indels need to be evaluated for non-random patterns not attributable to selection. The importance of nucleotide repeat patterns in the formation of indels should be further investigated. Repeat patterns should not be dismissed as evidence of degeneration in the genome, but need to be seriously evaluated as potential mechanisms for formatting the genome for future adaptive changes.
The observations of MC1R variation patterns in humans should be explored in more detail. At this point it appears that both the current evolutionist and creationist explanations of skin color variation are in need of an upgrade. Creationists need to recognize the existence of genes that were designed to handle mutations. It should be recognized that natural selection is being inferred based on statistical tests and the evolutionary assumption that mutations are essentially random accidents. Not only is differential reproduction not being demonstrated, but there is no plausible mechanism for sufficient selection to produce existing patterns. If natural selection cannot account for these non-random mutational patterns, then something else must be going on. This should provide creationists with strong motivation to pursue the possibility that directed mutations may often explain non-random patterns in mutations.
References
Anonymous. 1980. The origin of the human races. Ex nihilo 3(3):5–10.
Arendt, J. D. and D. N. Reznick. 2005. Evolution of juvenile growth rates in female guppies (Poecilia reticulata): Predator regime or resource level? Proceedings of the Royal Society B 272:333–337.
Bastiaens, M. T., J. A. ter Huurne, C. Kielich, N. A. Gruis, R. G. Westendorp, B. J. Vermeer, and J. N. Bavinck. 2001. Melanocortin-1 receptor gene variants determine the risk of nonmelanoma skin cancer independently of fair skin and red hair. American Journal of Human Genetics 68: 884–894.
Beaumont, K. A., S. N. Shekar, R. A. Newton, M. R. James, J. L. Stow, D. L. Duffy, and R. A. Sturm. 2007. Receptor function, dominant negative activity and phenotype correlations for MC1R variant alleles. Human Molecular Genetics 16:2249–2260.
Bennett, D. C. 2008. How to make a melanoma: what do we know of the primary clonal events? Pigment Cell and Melanoma Research 21:27–38.
Box, N. F., D. L. Duffy, R. E. Irving, A. Russell, W. Chen, L. R. Griffyths, P. G. Parsons, A. C. Green, and R. A. Sturm. 2001. Melanocortin-1 receptor genotype is a risk factor for basal and squamous cell carcinoma. Journal of Investigative Dermatology 116:224–229.
Byrne, K. and R. A. Nichols. 1999. Culex pipiens in London Underground tunnels: Differentiation between surface and subterranean populations. Heredity 82 (Pt 1):7–15.
Candille, S. I., C. B. Kaelin, B. M. Cattanach, B. Yu, D. A. Thompson, M. A. Nix, J. A. Kerns, S. M. Schmutz, G. L. Millhauser, and G. S. Barsh. 2007. A ß-defensin mutation causes black coat color in domestic dogs. Science 318: 1418–1423.
Eizirik, E., N. Yuhki, W. E. Johnson, M. Menotti-Raymond, S. S. Hannah, and S. J. O’Brien. 2003. Molecular genetics and evolution of melanism in the cat family. Current Biology 13:448–453.
Fargnoli, M. C., E. Altobelli, G. Keller, S. Chimenti, H. Höfler, and K. Peris. 2006. Contribution of melanocortin-1 receptor gene variants to sporadic cutaneous melanoma risk in a population in central Italy: A case-control study. Melanoma Research 16:175–182.
García-Borrón, J. C., B. L. Sánchez-Laorden, and C. Jiménez- Cervantes. 2005. Melanocortin-1 receptor structure and functional regulation. Pigment Cell Research 18:393–410.
Giuffra, E., J. M. H. Kijas, V. Amarger, Ö. Carlborg, J.-T. Jeon, and L. Andersson. 2000. The origin of the domestic pig: Independent domestication and subsequent introgression. Genetics 154:1785–1791.
Grant, P. R., and B. R. Grant. 2006. Evolution of character displacement in Darwin’s finches. Science 313(5784): 224–226.
Gustafsson, A. C., J. M. H. Kijas, A. Alderborn, M. Uhlén, L. Andersson, and J. Lundeberg. 2001. Screening and scanning of single nucleotide polymorphisms in the pig melanocortin 1 receptor gene (MC1R) by pyrosequencing. Animal Biotechnology 12(2):145–153.
Harding, R. M., E. Healy, A. J. Ray, N. S. Ellis, E. Flanagan, C. Todd, C. Dixon, A. Sajantila, I. J. Jackson, M. A. Birch- Machin, and J. L. Rees. 2000. Evidence for variable selective pressures at MC1R. American Journal of Human Genetics 66:1351–1361.
Herrel, A., K. Huyghe, B. Vanhooydonck, T. Backeljau, K. Breugelmans, I. Grbac, R. Van Damme, and D. J. Irschick. 2008. Rapid large-scale evolutionary divergence in morphology and performance associated with exploitation of a different dietary resource. Proceedings of the National Academy of Sciences of the United States of America 105(12):4792–4795.
Hosoda, T., J. J. Sato, T. Shimada, K. L. Campbell, and H. Suzuki. 2005. Independent nonframeshift deletions in the MC1R gene are not associated with melanistic coat coloration in three mustelid lineages. Journal of Heredity 96(5):607–613.
Kanetsky, P. A., F. Ge, D. Najarian, J. Swoyer, S. Panossian, L. Schuchter, R. Holmes, D. Guerry, and T. R. Rebbeck. 2004. Assessment of polymorphic variants in the melanocortin-1 receptor gene with cutaneous pigmentation using an evolutionary approach. Cancer Epidemiology, Biomarkers & Prevention 13(5):808–819.
Kennedy, C., J. ter Huurne, M. Berkhout, N. Gruis, M. Bastiaens, W. Bergman, R. Willemze, and J. N. Bavinck. 2001. Melanocortin 1 receptor (MC1R) gene variants are associated with an increased risk for cutaneous melanoma which is largely independent of skin type or hair color. Journal of Investigative Dermatology 117:294–300.
Kerns, J. A., E. J. Cargill, L. A. Clark, S. I. Candille, T. G. Berryere, M. Oliver, G. Lust, R. J. Todhunter, S. M. Schmutz, K. E. Murphy, and G. S. Barsh. 2007. Linkage and segregation analysis of black and brindle coat color in domestic dogs. Genetics 176:1679–1689.
Kijas, J. M. H., M. Moller, G. Plastow, and L. Andersson. 2001. A frameshift mutation in MC1R and a high frequency of somatic reversions cause black spotting in pigs. Genetics158:779–785.
Kijas, J. M. H., R. Wales, A. Törnsten, P. Chardon, M. Moller, and L. Andersson. 1998. Melanocortin receptor 1 (MC1R) mutations and coat color in pigs. Genetics 150:1177–1185.
Klungland, H., and D. I. Våge. 2000. Molecular genetics and pigmentation in domestic animals. Current Genomics 1(3):223–242.
Lachiewicz, A. M., M. Berwick, C. L. Wiggins, and N. E. Thomas. 2008. Epidemiologic support for melanoma heterogeneity using the surveillance, epidemiology, and end results program. Journal of Investigative Dermatology 128:1340–1342.
Moore, J. P. 1996. Skin deep. Creation Ex Nihilo 18(3):46–48.
Myers, P., R. Espinosa, C. S. Parr, T. Jones, G. S. Hammond, and T. A. Dewey. 2008. The animal diversity web (online). Retrieved June 30, 2008, from http://animaldiversity.org.
Newton, J. M., A. L. Wilkie, L. He, S. A. Jordan, D. L. Metallinos, N. G. Holmes, I. J. Jackson, and G. S. Barsh. 2000. Melanocortin 1 receptor variation in the domestic dog. Mammalian Genome 11:24–30.
Parker, G. 2006. Creation: Facts of life. Green Forest, Arkansas: Master Books.
Raimondi, S., F. Sera, S. Gandini, S. Iodice, S. Caini, P. Maisonneuve, and M. C. Fargnoli. 2008. MC1R variants, melanoma and red hair phenotype: A meta-analysis. International Journal of Cancer 122:2753–2760.
Ussher, J. 2003. The Annals of the World. Trans. and ed. L. and M. Pierce. Green Forest, Arkansas: Master Books.
Våge, D. I., M. R. Fleet, R. Ponz, R. T. Olsen, L. V. Monteagudo, M. T. Tejedor, M. V. Arruga, R. Gagliardi, A. Postiglioni, G. S. Nattrass, and H. Klungland. 2003. Mapping and characterization of the dominant black colour locus in sheep. Pigment Cell Research 16:693–697.
Våge, D. I., E. Fuglei, K. Snipstad, J. Beheim, V. M. Landsem, and H. Klungland. 2005. Two cysteine substitutions in the MC1R generate the blue variant of the arctic fox (Alopex lagopus) and prevent expression of the white winter coat. Peptides 26:1814–1817.
Våge, D. I., D. Lu, H. Klungland, S. Lien, S. Adalsteinsson, and R. D. Cone. 1997. A non-epistatic interaction of agouti and extension in the fox, Vulpes vulpes. Nature Genetics15:311–315.
Wada, A, T. Kunieda, M. Nishimura, Y. Kakizoe-Ishida, N. Watanabe, K. Ohkawa, and M. Tsudzuki. 2005. A nucleotide substitution responsible for the tawny coat color mutation carried by the MSKR inbred strain of mice. Journal of Heredity 96(2):145–149.
Walsh, J. S. 1999. Melanoma: Epidemiology and diagnosis. Jacksonville Medicine. Retrieved July 28, 2008, from http://www.dcmsonline.org/jax-medicine/1999journals/ Oct99/epi-diag.htm.
Wood, T. C. 2006. The current status of baraminology. Creation Research Society Quarterly 43:149–158.









